Increasing food intake by swapping mitochondrial genomes
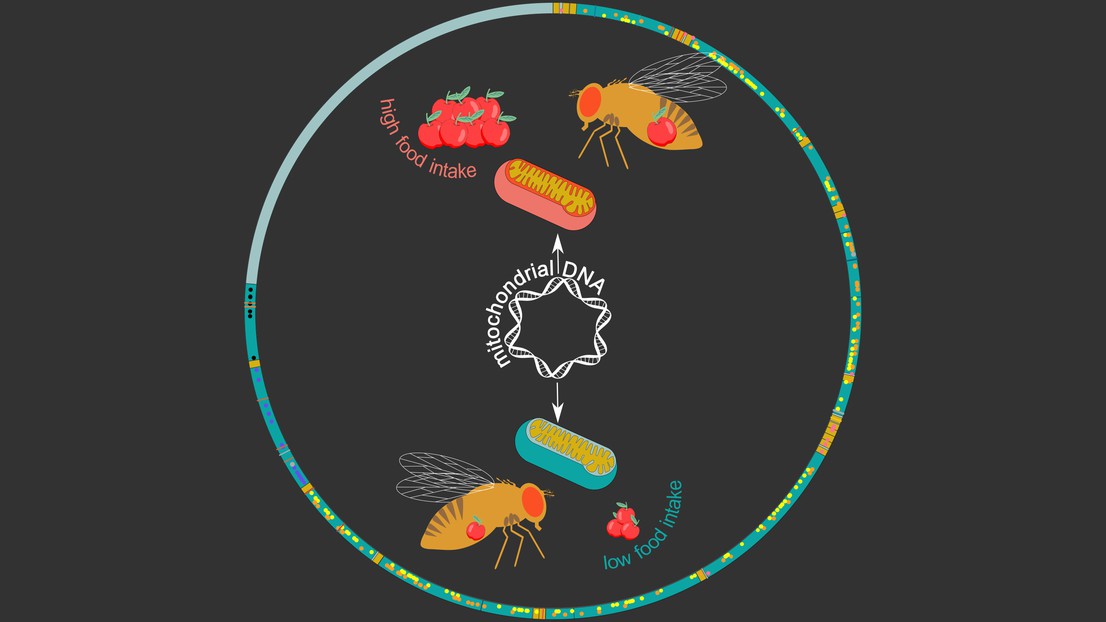
The Drosophila mitochondrial genome (circle). Credit: Maria Litovchenko/Roel Bevers
To uncover the relationship between variation in genes and phenotypic diversity, geneticists use a set of fully sequenced fruit-fly genomes. But little is known about the variation in the mitochondrial genome, for which mutations are linked to an array of diseases. Now, EPFL scientists have created a high-resolution map of mitochondrial DNA variants in the fruit fly, connecting mitochondrial genes to metabolic traits and diseases.
Image: The Drosophila mitochondrial genome (circle) with genes in dark green and dots/lines depicting genetic variants. The inner image shows two mitochondrial haplotypes: green linked to low food intake and red to high food intake. Credit: Maria Litovchenko/Roel Bevers
Many of the characteristics that make people so different from each other, are often the result of small differences in the DNA between individuals. Variation in just a single base in our DNA can cause significant variation in traits like sleep length, weight, height etc. And, unfortunately, they can also become a source of disease.
To confidently connect variation within a gene or genome (what biologists call “genotypes”) and variation in traits (“phenotypes”), scientists ideally need standardized genetic tools. One of these is the Drosophila Genetic Reference Panel (DGRP), which consists of more than 200 lines of the fruit fly Drosophila melanogaster. Each line in the DGRP has had its genome fully sequenced so that scientists can spot differences between genotypes and then connect these to differences between phenotypes of interest.
Most of the DNA in eukaryotic cells – the cells that make up higher organisms like fruit flies and humans – sits tightly packed inside the cell’s nucleus. But almost all eukaryotic cells also have mitochondria, an organelle often referred to as the “powerhouse of the cell”. Mitochondria are unique in that they have their own DNA with its own distinct set of genes.
Studies have hinted that variation in mitochondrial genes is associated with diseases like obesity, type-2 diabetes, multiple sclerosis, and schizophrenia. The problem is that, unlike “regular” genes in nuclear genomes, we actually know very little about how variation in mitochondrial genes relates to variation in phenotypes.
This is what Roel Bevers and Maria Litovchenko from the lab of Bart Deplancke at EPFL have addressed in a paper published in Nature Metabolism. The researchers carried out a comprehensive study connecting variation in mitochondrial DNA with various traits in DGRP fruit-fly lines. The findings can be incorporated into future genome-wide association studies (GWAS) to gain more detailed information about how variation in genes – nuclear and mitochondrial, and their interactions – affect various traits and diseases.
“So far, DGRP studies have mostly focused on genetic variants in the nucleus because of low mitochondrial DNA coverage,” says Deplancke. “In this study, we wanted to investigate the impact of mitochondrial DNA variation on traits in these fly lines.”
His team re-sequenced mitochondrial genomes of 169 DGRP fruit-fly lines, which allowed them to identify 231 gene variants. They then looked at “mitochondrial DNA haplotypes”, which are groups of variants which are always inherited together, identifying 12 distinct haplotypes among the studied fly lines.
Interestingly, these haplotypes were found to be associated with a range of metabolic phenotypes and diseases. “We found many links to stress- and metabolism-related traits, including food intake in males,” says Deplancke. To confirm this, the researchers swapped mitochondrial genomes between fruit fly lines, which actually changed the eating behavior of males: the mitochondrial haplotype associated with high food intake could increase food intake in fly lines that initially displayed a low food-intake.
The work can help future studies by providing a higher resolution to connect genes and traits, e.g. feeding behavior. “There is important mitochondrial haplotype-specific metabolic variation in the DGRP,” conclude the authors. “This demonstrates the importance of incorporating such haplotypes in genotype-phenotype relationship studies.”
Other contributors
- EPFL (Laboratory of Integrative Systems Physiology)
- Swiss Institute of Bioinformatics
- University of Lausanne (Complex Trait Genetics Group, Robinson Lab)
The Swiss Initiative in Systems Biology SystemsX.ch (AgingX project), EPFL
Roel P.J. Bevers & Maria Litovchenko, Adamandia Kapopoulou, Virginie S. Braman, Matthew R. Robinson, Johan Auwerx, Brian Hollis, Bart Deplancke. Mitochondrial haplotypes affect metabolic phenotypes in the Drosophila Genetic Reference Panel. Nature Metabolism 09 December 2019. DOI: 10.1038/s42255-019-0147-3